-
- General description
-
We have performed an extensive set of experiments to evaluate the effectiveness and efficiency of our approach on a number of different real and synthetic datasets.
Table. 1: Experimental data.Dataset Type Number of graphs Size on disk Average size DS1 Synthetic 20,000 18 MB [50-100] DS2 Synthetic 100,000 81 MB [50-70] DS3 Real 274,860 97 MB [40-50] DS4 Synthetic 500,000 402 MB [60-70] DS5 Synthetic 1,500,000 1.2 GB [60-70] DS6 Synthetic 100,000,000 69 GB [20-100]
The synthetic datasets are generated by the synthetic data generator GraphGen provided by Kuramochi and Karypis [1].
The real dataset we tested is a chemical compound dataset.
[1] M. Kuramochi, G. Karypis. Frequent Subgraph Discovery. In Proceedings of the 2001 IEEE International Conference on Data Mining, ICDM '01. IEEE Computer Society, Washington, DC, USA, 2001, pp. 313-320.
- Data sources
- The real dataset we tested is available from the Developmental Therapeutics Program (DTP) web site at National Cancer Institute (NCI). http://dtp.nci.nih.gov/branches/npb/repository.html
- GraphGen tool was used to generate our synthetic datasets. Download GraphGen
-
We implemented our approach in Perl language and we used Hadoop, (version 0.20.1), an open source version of MapReduce.
All the experiments of our approach were carried out using a local cluster with five nodes.
The processing nodes used in our tests are equipped with a Quad-Core AMD Opteron(TM) Processor 6234 2.40 GHz CPU and 4 GB of memory for each node.
- Effect of the partitioning method on the rate of lost subgraphs
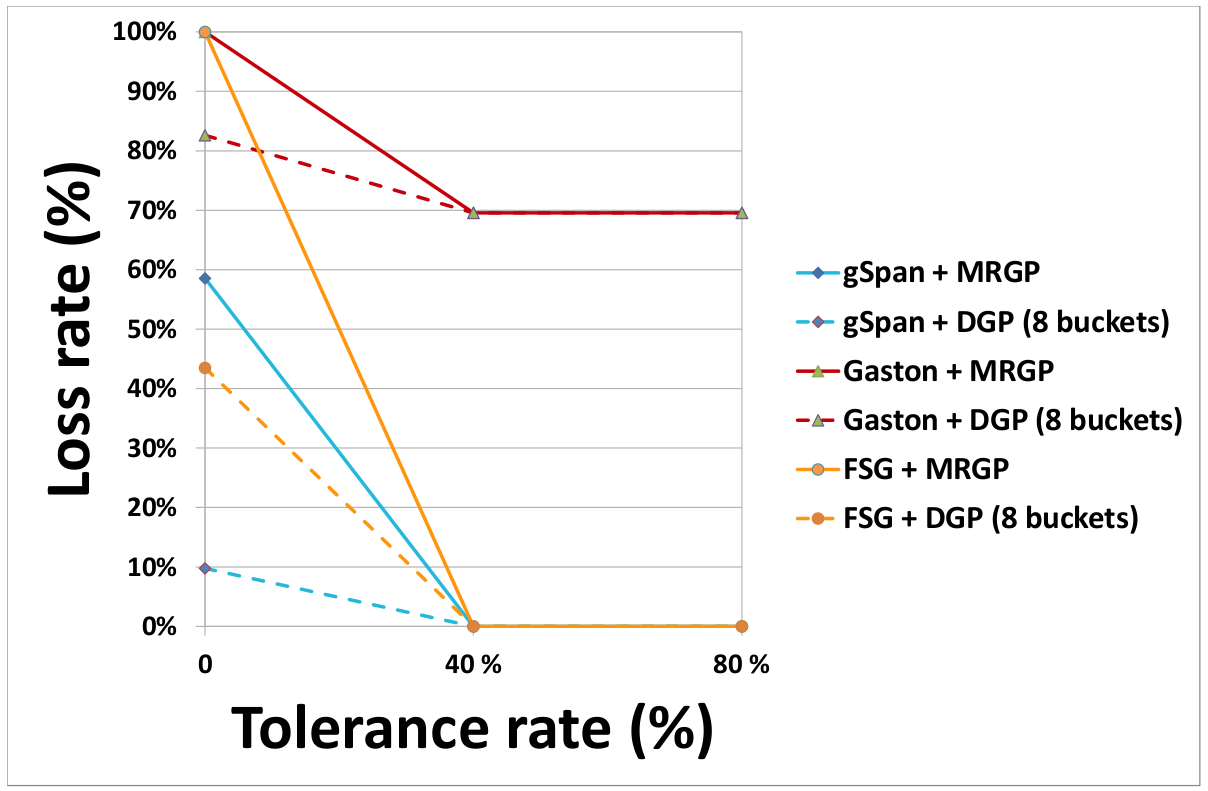
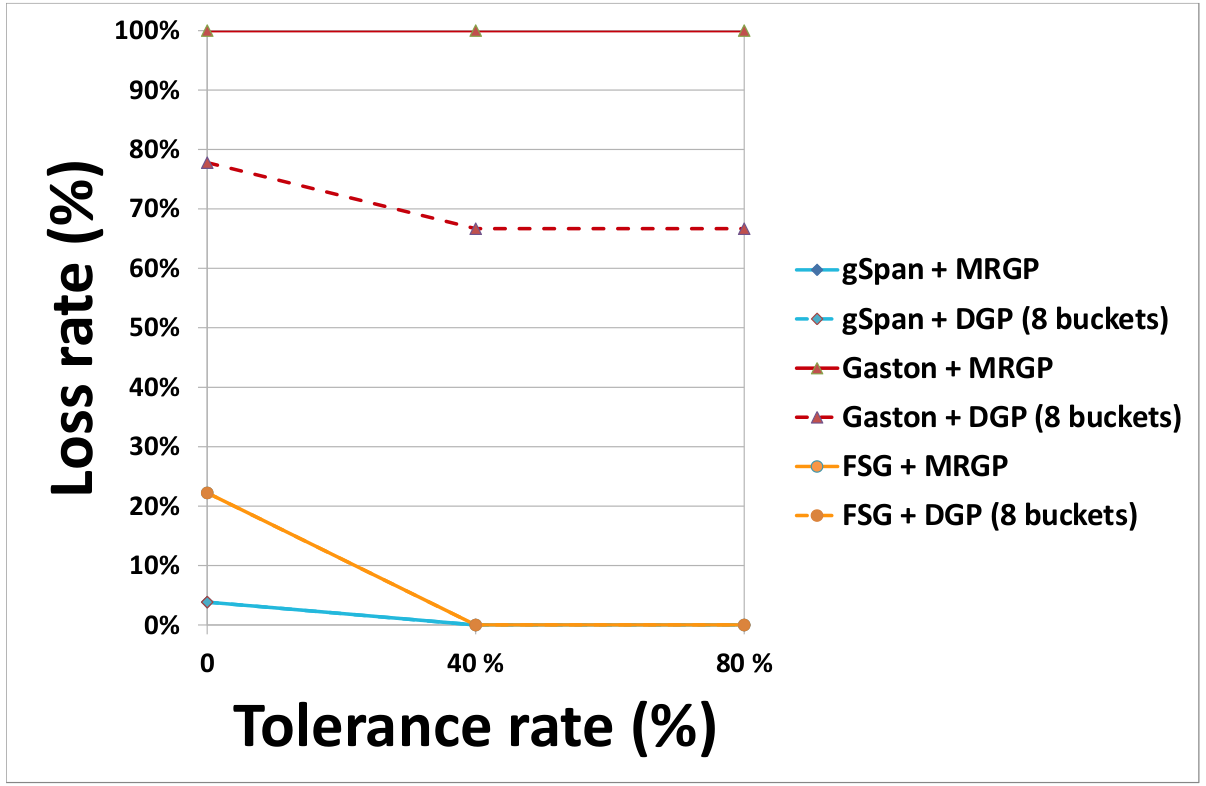
(a) DS1 (b) DS2 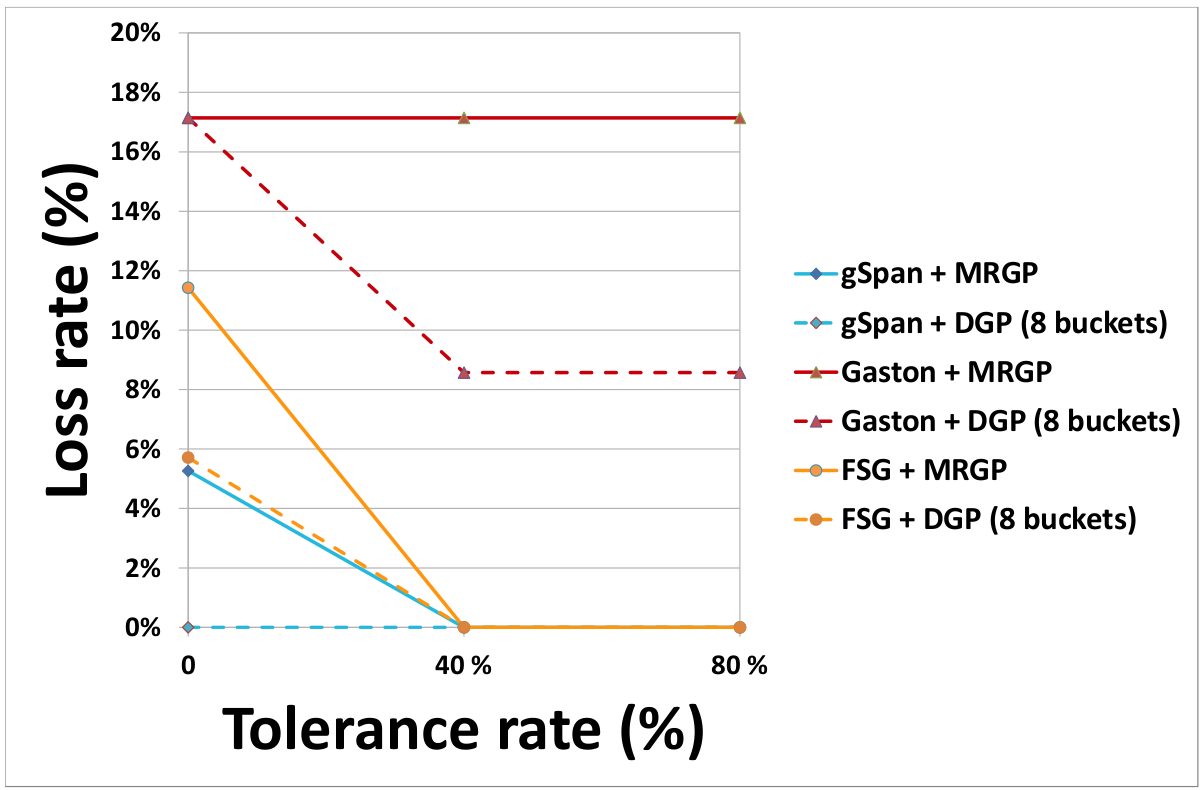
(c) DS3 - Speedup and Scalability
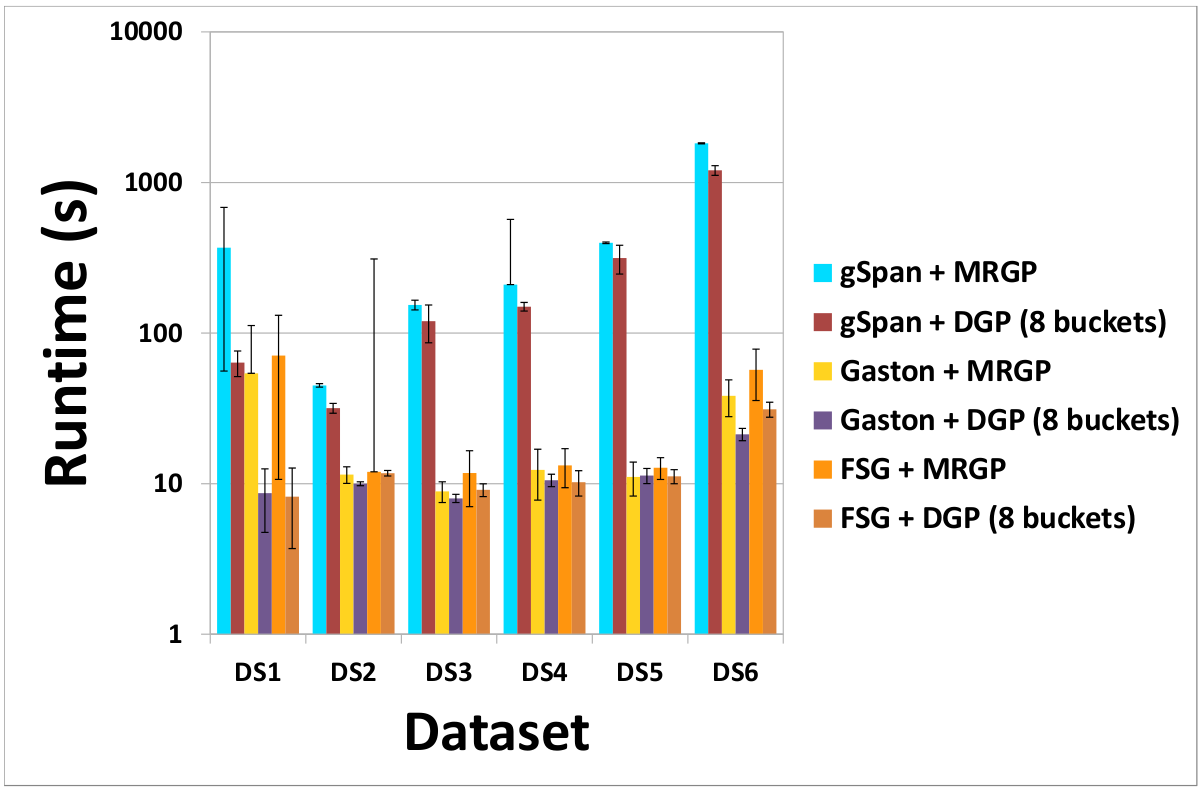
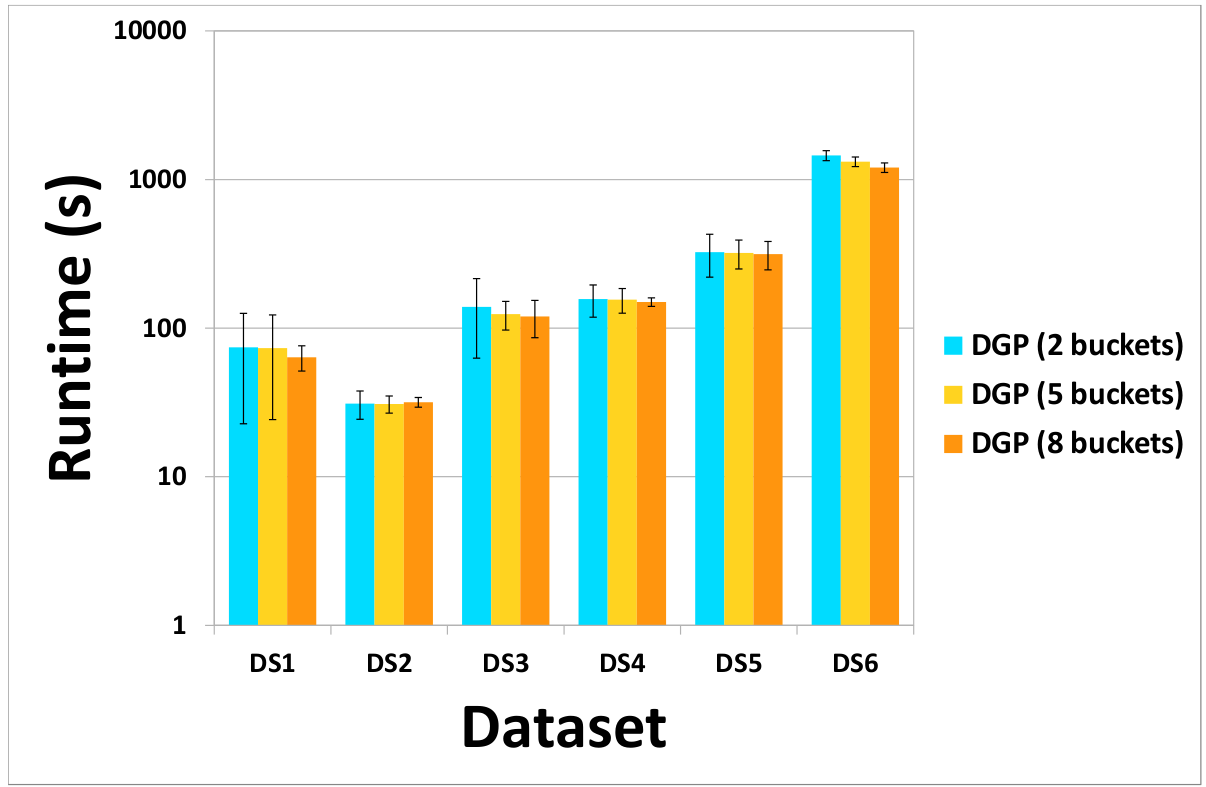
(a) Cost of DGP and MRGP partitioning methods (b) Effect of the number of buckets on the cost of the DGP method 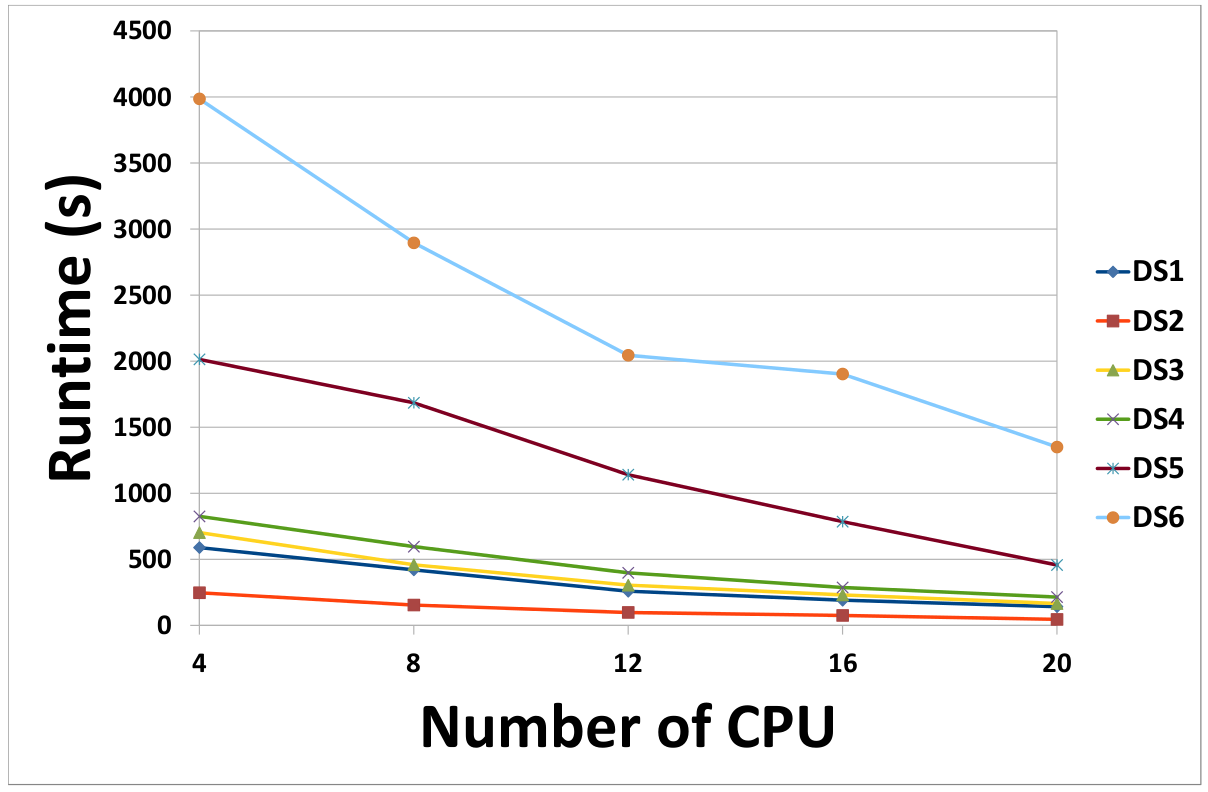
(c) Effect of the number of workers on the runtime of our MapReduce-based framework - Chunk size and replication factor
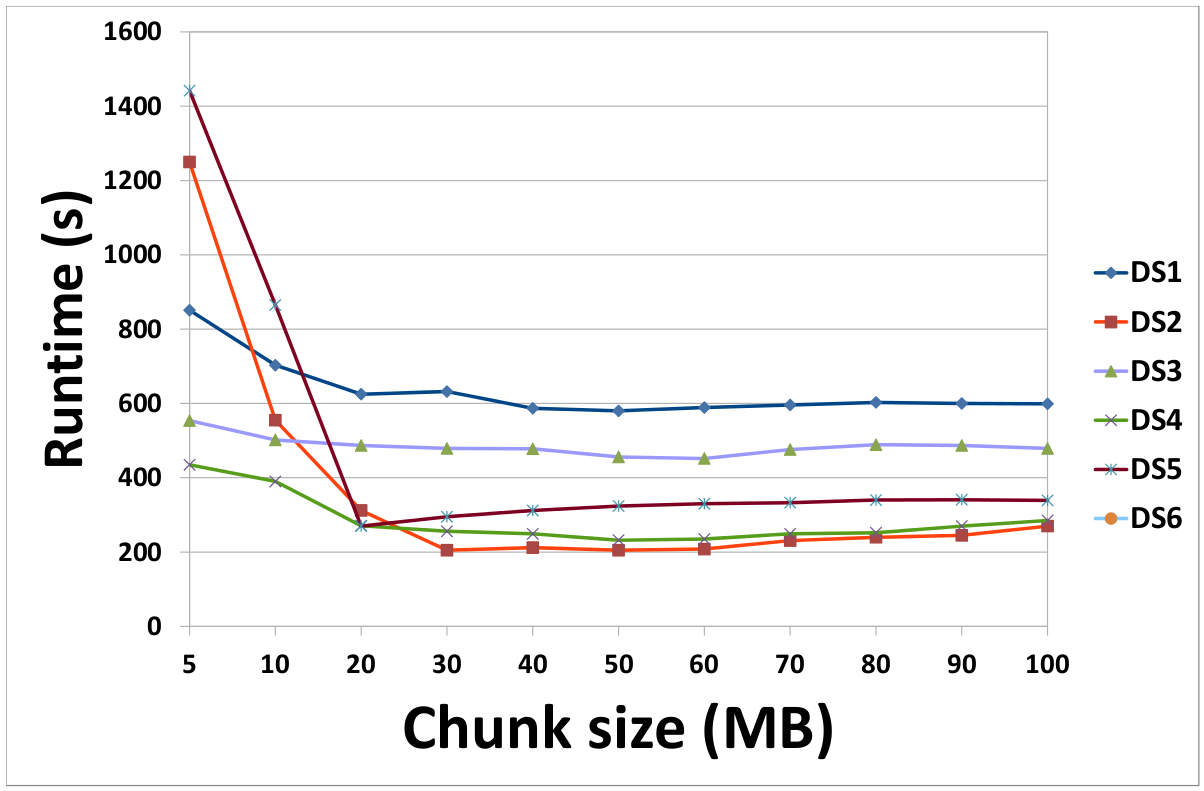
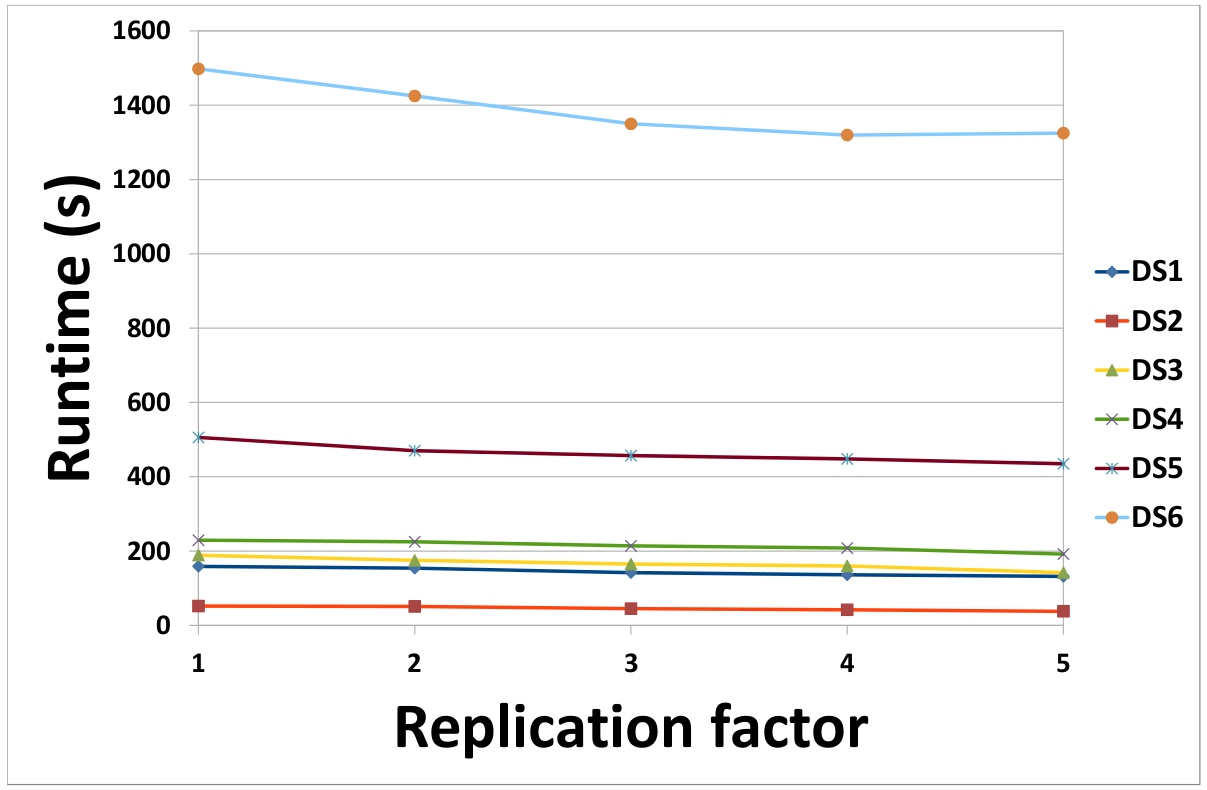
(a) Effect of chunk size on the runtime of our MapReduce-based framework (b) Effect of the number of copies of data on the runtime of our MapReduce-based framework
-
An implementation of our DGP (Density-based partitioning) method is available here.
A Hadoop implementation of our MapReduce-based framework for distributed frequent subgraph mining is available here.
The used subgraph extractors can be found here.
Instructions describing how to setup a Hadoop cluster on Ubuntu 14.04 can be found here. You can get a PDF copy from here.