3D modeling of protein – glycosaminoglycans (GAG) complexes
Collaboration Sergey Samsonov (Gdansk University)
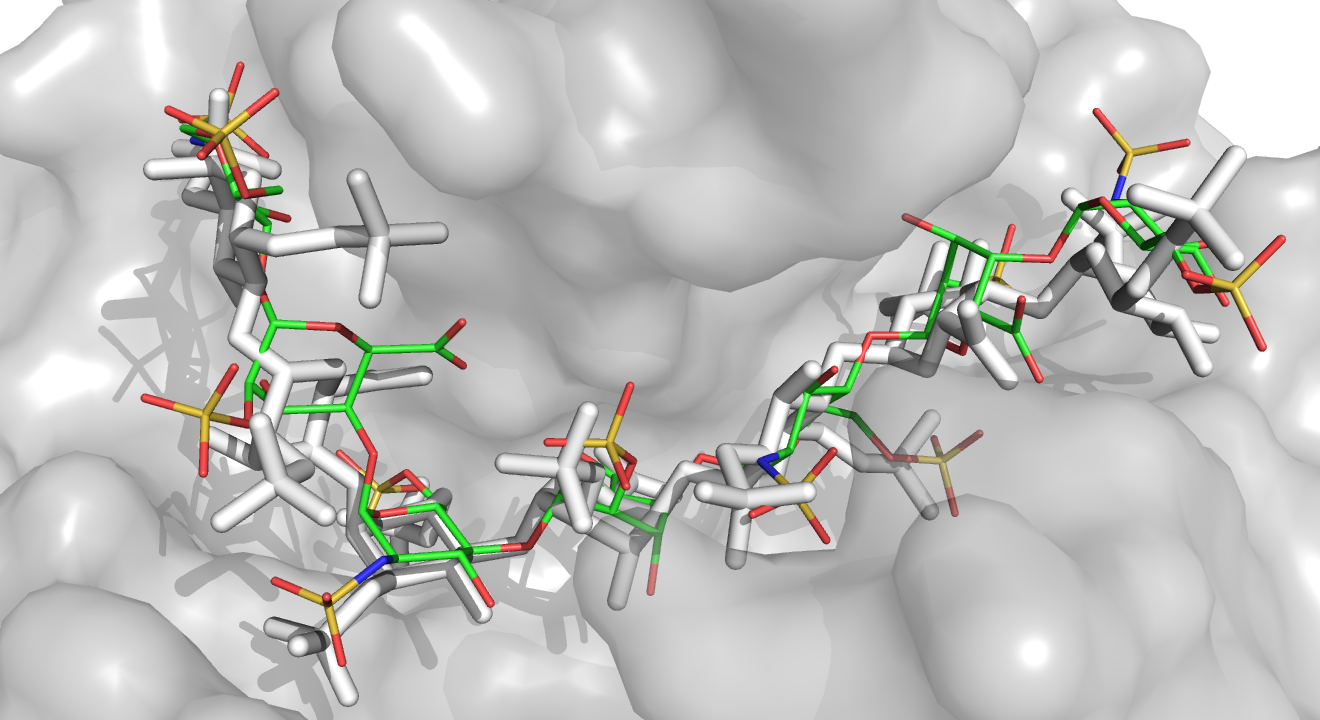
Glycosaminoglycans (GAGs) are periodic anionic linear polysaccharides that bind protein targets in the extracellular matrix. These are very promising biological objects for the design of new functional biomaterials, with medical applications such as regeneration of bones or skin [Scharnweber et al., JMSM 2015].
Useful biological and therapeutic knowledge can be drawn from the structure of complexes between GAGs and their target proteins. However, as for protein-ssRNA complexes, the experimental resolution as well as the modeling of their structure are very difficult. The reasons for this are both the intrinsic properties of GAGs (high flexibility and conformational diversity, high anionic charge) and the lack of specially designed computational tools for GAG-protein systems. This currently limits the docking of GAGs to very short fragments (~ tetramers) [Samsonov and Pisabarro, Glycobiology 2016].
We are developing new automated fragment docking methods of GAG on a protein, by combination of flexible fragment docking w. AutoDock and combinatorial assembly of compatible poses. This work contributes to enriching a still limited pool of computational tools specially developed for protein-GAG complexes.



